
Top left: reconstruction error for each dimensionality reduction method... | Download Scientific Diagram
![PDF] Efficient Model Selection for Mixtures of Probabilistic PCA Via Hierarchical BIC | Semantic Scholar PDF] Efficient Model Selection for Mixtures of Probabilistic PCA Via Hierarchical BIC | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/d7267bc9049cb1e5ba30f672424c35456b6c7771/8-Figure2-1.png)
PDF] Efficient Model Selection for Mixtures of Probabilistic PCA Via Hierarchical BIC | Semantic Scholar
PLOS ONE: Classification of cannabis strains in the Canadian market with discriminant analysis of principal components using genome-wide single nucleotide polymorphisms

Poisson PCA: Poisson measurement error corrected PCA, with application to microbiome data - Kenney - - Biometrics - Wiley Online Library

BIC statistics as a function of the number of knots for linear (solid... | Download Scientific Diagram

Model selection techniques for sparse weight‐based principal component analysis - Schipper - 2021 - Journal of Chemometrics - Wiley Online Library
Contour plot of BIC as a function of sumabsu and sumabsv for the first... | Download Scientific Diagram
![PDF] Efficient Model Selection for Mixtures of Probabilistic PCA Via Hierarchical BIC | Semantic Scholar PDF] Efficient Model Selection for Mixtures of Probabilistic PCA Via Hierarchical BIC | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/d7267bc9049cb1e5ba30f672424c35456b6c7771/9-Figure3-1.png)
PDF] Efficient Model Selection for Mixtures of Probabilistic PCA Via Hierarchical BIC | Semantic Scholar
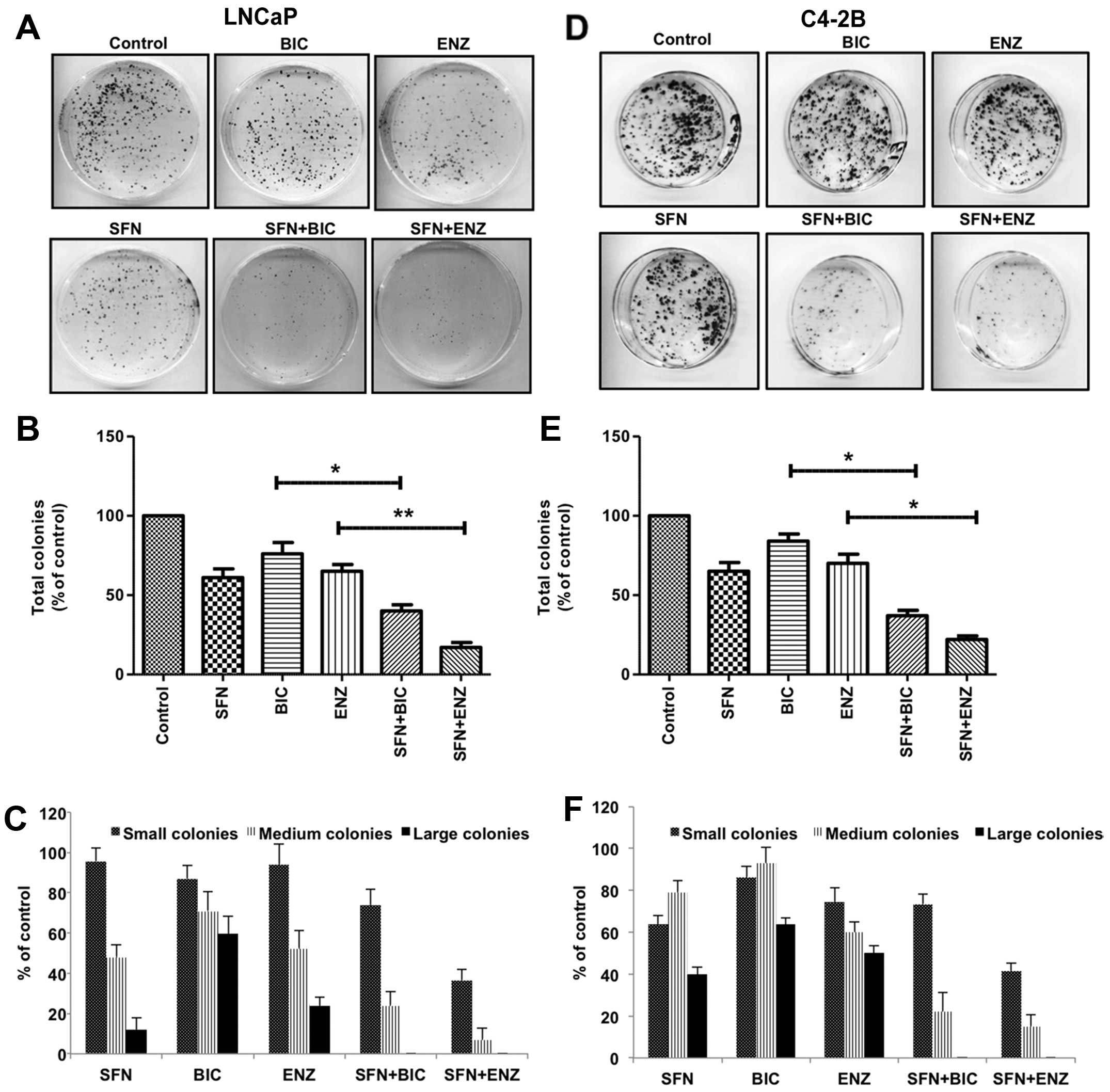
Sulforaphane increases the efficacy of anti-androgens by rapidly decreasing androgen receptor levels in prostate cancer cells

When using the find.clusters function in adegenet (DAPC), can the lowest BIC value be considered as an optimal BIC if this value is lower than 0?



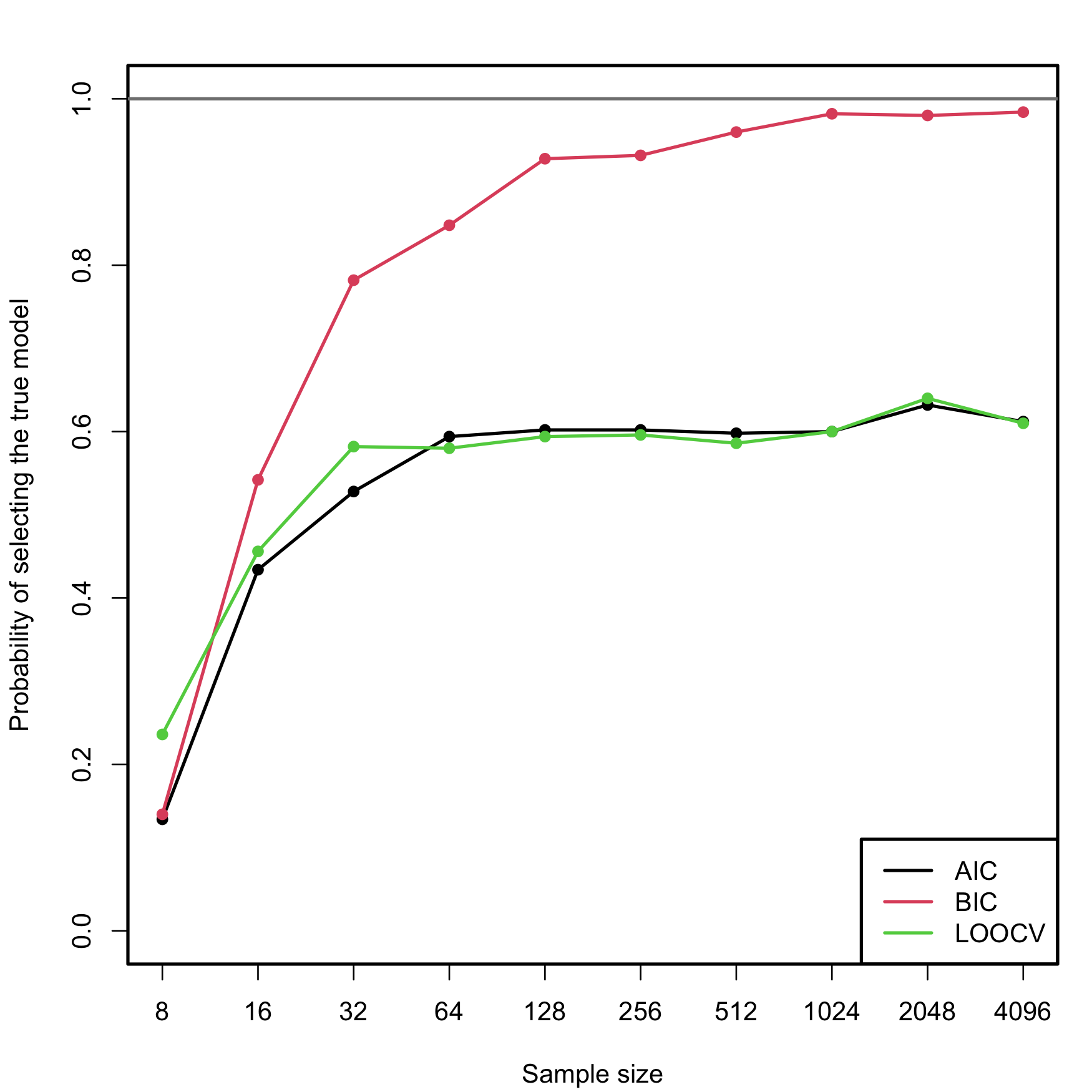
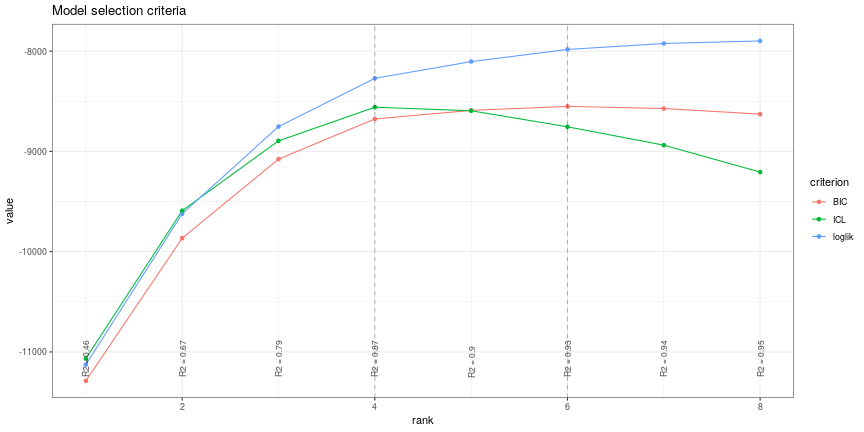
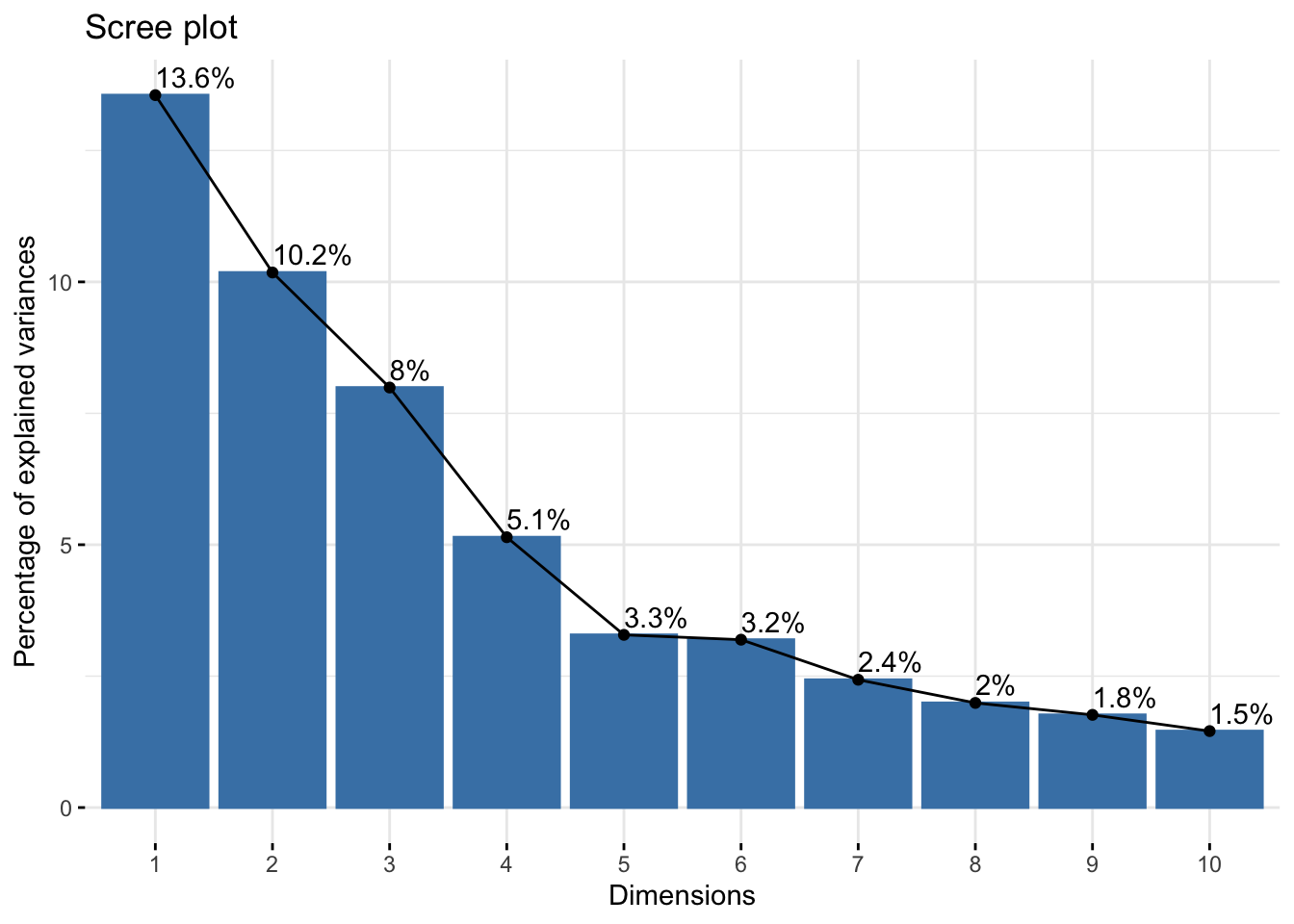

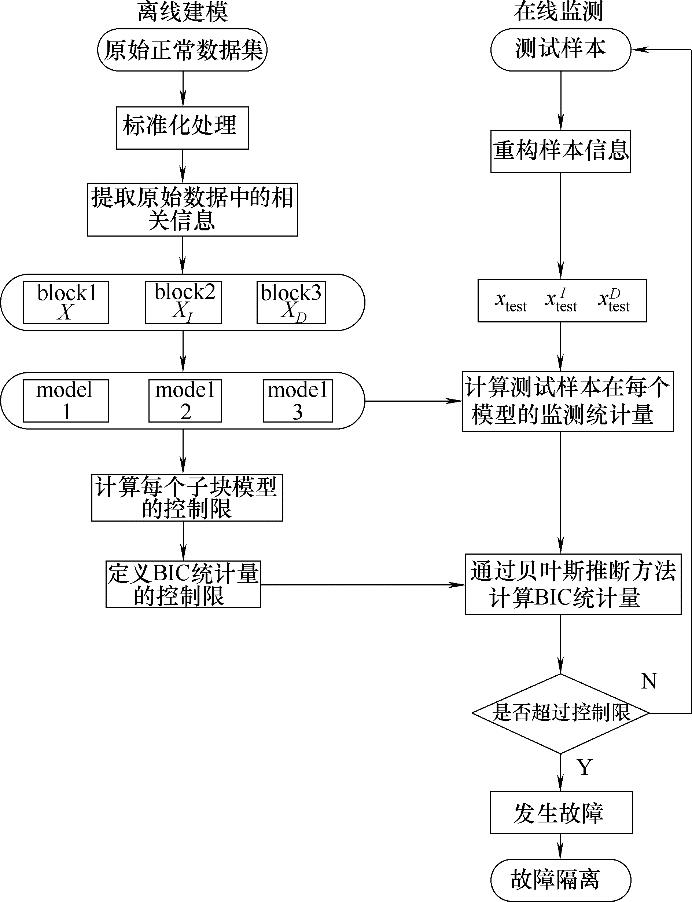

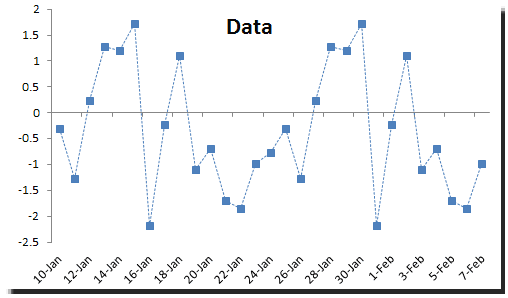

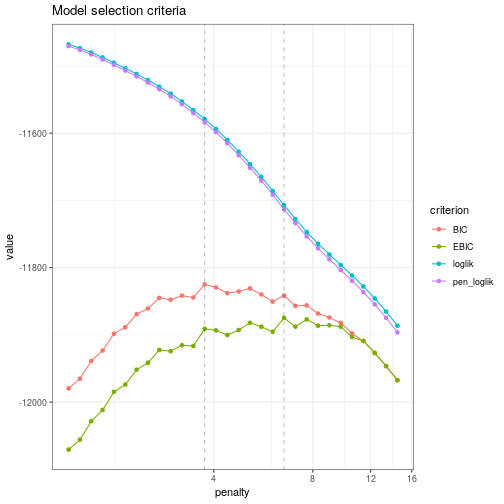

![PDF] Sparse variable noisy PCA using l0 penalty | Semantic Scholar PDF] Sparse variable noisy PCA using l0 penalty | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/bf51a128d4e13a92f6f3c309af08ed11e3a0ccaa/4-Figure3-1.png)


